
MalariaGEN is built on the idea that genomic data – ideally lots of genomic data – will deepen our understanding of malaria in profound ways. Recent research, led by Professor Olivo Miotto, has yielded a discovery that would not have been possible without the vast open-source datasets MalariaGEN produces.
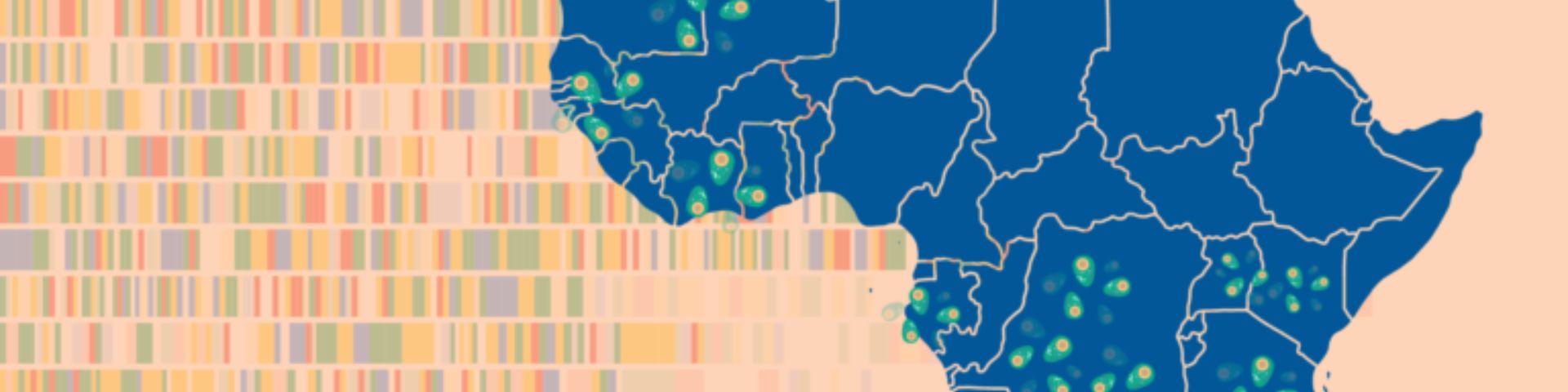
Researchers found a previously unknown group of related Plasmodium falciparum parasites, dubbed AF1, scattered thinly across the African continent.
This “strongly differentiated cluster of parasites” is remarkable for the number of the same genetic variations they share – up to 23 across the parasite’s genome – as well as their vast geographical range and relative scarcity wherever they do appear.
The related group was detected in 13 countries, stretching from Mauritania to Madagascar – a range of over 7,000 kilometres. And yet on average, AF1 parasites accounted for just 1.2% of the samples analysed.
This is exactly the kind of faint pattern that could only be observed by studying large datasets. Scientists sifted through 4,376 African parasite genomes from MalariaGEN’s Pf7 dataset – the largest global collection of P. falciparum data with over 20,000 genome sequences across 33 countries.
Because the rarity of these parasites makes them hard to detect, researchers have coined the term cryptotype. “It’s not a strain of parasites you can see at first glance – you have to do some rather intricate analysis to find it,” explains Professor Miotto. “None of this could have been done without the MalariaGEN whole-genome dataset. It’s an immensely useful and important tool.”
Unlike bacteria or viruses, malaria parasites reproduce sexually. That means the offspring inherits genetic material from both parents.
As a result, complex combinations of variants – such as those in AF1 – are quickly lost from a population. The chances of sustaining 23 different rare variations, spread across nearly all the parasite’s chromosomes, is highly unlikely. Unless something unusual is going on.
Distinct groups do occur in certain circumstances, for example in locations where malaria transmission is low. Plasmodium reproduces in a mosquito’s midgut, but if that mosquito is only being infected by parasites from one human then clones of the same parasite end up reproducing with each other. This is called “selfing”. The same might happen in parasites which have specialised to infect a particular species of mosquito that isn’t very common in that environment.
That isn’t happening with AF1. In fact, with the exception of those shared regions, the genomic analysis shows these parasites are also related to – and therefore breeding with – the non-AF1 parasites living nearby. And yet the collection of rare mutations persists, in low numbers, right across Africa.
A possible answer, the researchers propose, is that this “complex compendium of interacting variants” has allowed the AF1 parasites to “adapt to some as-yet-unidentified evolutionary niche” and given them some sort of selective advantage which means these genetic variations are more likely to be passed on to the next generation.
Genetic variants often need to occur together to provide an evolutionary advantage. A variant on its own may give an advantage in one way, but disadvantage the parasite in others. This disadvantage may then be compensated for by another variant elsewhere in the genome. This is likely to be the case with AF1, since the shared variants are rarely seen by themselves in other parasites, but have existed together in the AF1 group over a long period of time and spread to many places.
It is therefore likely these variants are working together, presumably to give the AF1 parasites an evolutionary advantage under certain conditions. This condition may not be very common – remember, AF1 parasites were only 1.2% of samples – but it happens enough, and it happens right across Africa: from the Gambia to Gabon, Mali to Malawi.
The specific advantage remains unclear. But it is interesting that many of these variants involve genes linked to how parasites invade and then exit human red blood cells. Their low numbers have led to suggestions that the AF1 parasites may have found an advantage when interacting with the red blood cells of certain people.
Now that we know these AF1 parasites exist, scientists can begin asking why. The suggestion that their genetic variations help the parasites interact with red blood cells can be explored. High-quality long-read genomic sequencing of AF1 parasites can provide further insights into the group. Meanwhile, identifying patients infected by AF1 parasites may help shed more light on their evolutionary niche.
Such investigations could be challenging for the simple fact that there aren’t many AF1 parasites around. However, understanding this group may improve our understanding of the invasive mechanisms used by P. falciparum parasites more generally.
This result is also proof of principle that there may also be other cryptotypes waiting to be discovered.
“We’re sitting on a huge treasure trove of data,” says Professor Miotto. “I think if this study proves anything, it’s that you can take this [dataset], slice it in any way, and find amazing things.”